#4 mRNA and protein modules regulated by IFN-α
Mireia Ramos-Rodríguez
Details
Original publication:
Colli, M.L., Ramos-Rodríguez, M., Nakayasu, E.S. et al. An integrated multi-omics approach identifies the landscape of interferon-α-mediated responses of human pancreatic beta cells. Nat Commun 11, 2584 (2020). https://doi.org/10.1038/s41467-020-16327-0
Contents: Analyses and figures contained in this document correspond to the following figures/sections of the original publication:
- Results: “mRNA and protein modules regulated by interferon-\(\alpha\)”.
- Figure 4: Weighted correlation network analysis (WGCNA) identifies IFN\(\alpha\)-regulated mRNA and protein modules. Panels d and e.
Open chromatin in mRNA–protein modules
Association of chromatin accessibility with mRNA–protein modules
ATAC-seq-identified open chromatin regions were linked to gene transcription start sites (TSSs) in a 40 kb window. These genes and their open chromatin regions were associated to the modules of DEG and DAP.
load("../data/IFNa/clusters_RNAProt/gene-protein_granges.rda")
load("../data/IFNa/ATAC/diffAnalysis/res_2h_granges.rda")
res.2h.gr <- res.gr
load("../data/IFNa/ATAC/diffAnalysis/res_24h_granges.rda")
res.24h.gr <- res.gr
## Annotate to 40kb window
win <- 40000
gp.prom <- promoters(gp, upstream=win/2, downstream=win/2)
hits.2h <- findOverlaps(res.2h.gr, gp.prom)
hits.24h <- findOverlaps(res.24h.gr, gp.prom)
anno.2h <- cbind(data.frame(res.2h.gr)[queryHits(hits.2h),c(6,8,12:13)],
data.frame(gp.prom)[subjectHits(hits.2h),c(6:11)])
anno.24h <- cbind(data.frame(res.24h.gr)[queryHits(hits.24h),c(6,8,12:13)],
data.frame(gp.prom)[subjectHits(hits.24h),c(6:11)])
## Melt data.frames
load("../data/IFNa/ATAC/diffAnalysis/res.2h.rda")
load("../data/IFNa/ATAC/diffAnalysis/res.24h.rda")
anno.2h <- dplyr::left_join(anno.2h, res.2h.df[,c(1,8:15)])
anno.2h$log2FoldChange[is.na(anno.2h$log2FoldChange)] <- 0
anno.2h$mean.ctrl <- unlist(apply(anno.2h[,11:14], 1, mean))
anno.2h$mean.ifn <- unlist(apply(anno.2h[,15:18], 1, mean))
anno.2h <- anno.2h[,c(1:10,19:20)]
anno.2h.m <- reshape2::melt(anno.2h,
id.vars=1:10,
value.vars=11:12,
value.name="counts",
variable.name="treatment")
anno.2h.m$treatment <- gsub("mean.", "", anno.2h.m$treatment)
anno.24h <- dplyr::left_join(anno.24h, res.24h.df[,c(1,8:15)])
anno.24h$log2FoldChange[is.na(anno.24h$log2FoldChange)] <- 0
anno.24h$mean.ctrl <- unlist(apply(anno.24h[,11:14], 1, mean))
anno.24h$mean.ifn <- unlist(apply(anno.24h[,15:18], 1, mean))
anno.24h <- anno.24h[,c(1:10,19:20)]
anno.24h.m <- reshape2::melt(anno.24h,
id.vars=1:10,
value.vars=11:12,
value.name="counts",
variable.name="treatment")
anno.24h.m$treatment <- gsub("mean.", "", anno.24h.m$treatment)The enrichment for gained open chromatin regions was then evaluated using Chi-squared tests.
files <- list.files("../data/IFNa/clusters_RNAProt/",
pattern="^cluster*", full.names=TRUE)
mod <- lapply(files, read.csv, stringsAsFactors=F)
names <- pipelineNGS::getNameFromPath(files, suffix=".csv", prefix="cluster")
mod.comb <- do.call(rbind, mod)
mod.comb$cluster <- unlist(mapply(rep, names, each=sapply(mod, nrow)))
colnames(mod.comb)[1] <- "external_gene_name"
anno.2h <- dplyr::left_join(anno.2h, mod.comb[c(1,6)])
chisq.test(table(anno.2h$cluster, anno.2h$type))
Pearson's Chi-squared test
data: table(anno.2h$cluster, anno.2h$type)
X-squared = 15.583, df = 4, p-value = 0.003633ggplot(anno.2h[!is.na(anno.2h$cluster),],
aes(cluster, ..count.., fill=type)) +
scale_fill_manual(values=c("gained"="dark green",
"lost"="dark red",
"stable"="grey"),
name="Chromatin Type") +
geom_bar(lwd=0.7, color="black") +
ylab("# Regions") +
theme(legend.position="top",
axis.text.x=element_text(angle=30, hjust=1))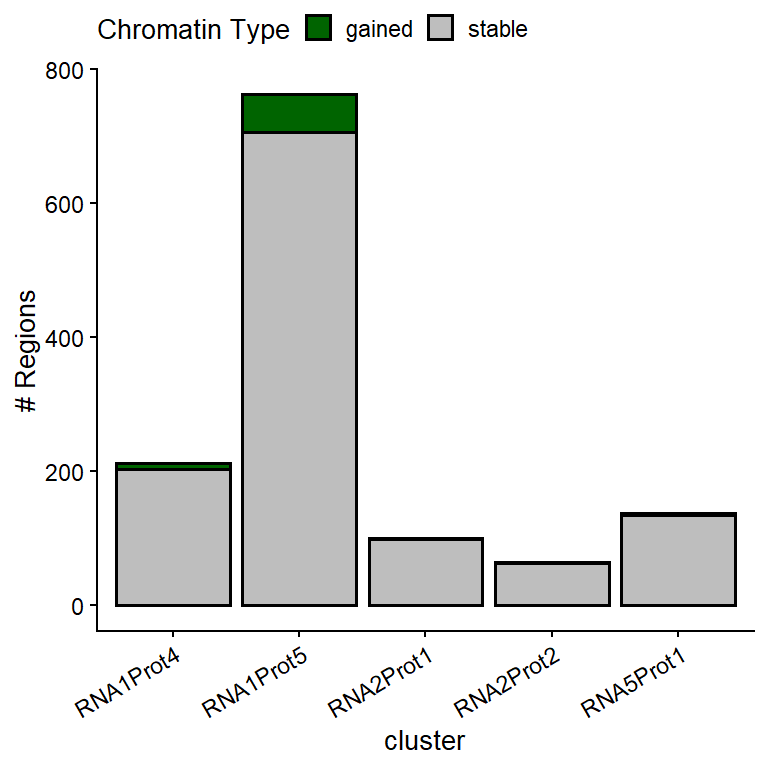
Figure 1: Type of OCRs associated to each RNA-Prot module.
Characterization of TF motifs found in module #2
As module #2 (Rna1Prot5) showed significant enrichment of gained OCRs, a de novo motif analysis was performed to determine the TF motifs present in such ATAC-seq regions.
library(maRge)
deNovoMotifHOMER(bed="gainedATAC_RNA1Prot5.bed",
path_output=file.path(out_dir, "motifs_gainedATAC-RNA1Prot5"),
path_homer="~/tools/homer/")htmltools::includeHTML(file.path(out_dir, "motifs_gainedATAC-RNA1Prot5/homerResults.html"))Homer de novo Motif Results (motifs_gainedATAC-RNA1Prot5/)
Known Motif Enrichment ResultsGene Ontology Enrichment Results
If Homer is having trouble matching a motif to a known motif, try copy/pasting the matrix file into STAMP
More information on motif finding results: HOMER | Description of Results | Tips
Total target sequences = 55
Total background sequences = 49704
* - possible false positive
| Rank | Motif | P-value | log P-pvalue | % of Targets | % of Background | STD(Bg STD) | Best Match/Details | Motif File |
| 1 | 1e-26 | -6.082e+01 | 45.45% | 2.16% | 114.3bp (175.5bp) | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer(0.959) More Information | Similar Motifs Found | motif file (matrix) | |
| 2 * | 1e-8 | -1.847e+01 | 23.64% | 3.08% | 255.1bp (176.7bp) | Egr2(Zf)/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer(0.666) More Information | Similar Motifs Found | motif file (matrix) | |
| 3 * | 1e-6 | -1.499e+01 | 27.27% | 5.73% | 215.6bp (177.4bp) | REL/MA0101.1/Jaspar(0.786) More Information | Similar Motifs Found | motif file (matrix) | |
| 4 * | 1e-5 | -1.286e+01 | 27.27% | 6.79% | 175.5bp (181.2bp) | Ascl2/MA0816.1/Jaspar(0.741) More Information | Similar Motifs Found | motif file (matrix) | |
| 5 * | 1e-4 | -1.094e+01 | 27.27% | 7.96% | 292.8bp (177.2bp) | HMBOX1/MA0895.1/Jaspar(0.865) More Information | Similar Motifs Found | motif file (matrix) | |
| 6 * | 1e-4 | -1.052e+01 | 7.27% | 0.31% | 50.2bp (168.1bp) | PB0137.1_Irf3_2/Jaspar(0.598) More Information | Similar Motifs Found | motif file (matrix) | |
| 7 * | 1e-4 | -1.015e+01 | 18.18% | 3.81% | 161.2bp (175.0bp) | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer(0.733) More Information | Similar Motifs Found | motif file (matrix) | |
| 8 * | 1e-2 | -6.114e+00 | 1.82% | 0.01% | 17.1bp (116.8bp) | PB0110.1_Bcl6b_2/Jaspar(0.629) More Information | Similar Motifs Found | motif file (matrix) | |
| 9 * | 1e-2 | -5.656e+00 | 14.55% | 4.59% | 122.1bp (171.2bp) | PB0138.1_Irf4_2/Jaspar(0.667) More Information | Similar Motifs Found | motif file (matrix) |